Group Burgering
Prof.Dr. Boudewijn Burgering Professor of Signal Transduction
Telephone number: + 31 88 7568918 /8988
E-mail address: B.M.T.Burgering@umcutrecht.nl

Research program / group uitklapper, klik om te openen
The PI3K/PKB(AKT)/FOXO signaling pathway
The signaling emanating from the lipid kinase PI3K is involved in a number of interesting and important cellular and organismal functions. In insulin signaling PI3K signaling drives important metabolic features and is the major pathway deregulated in insulin-dependent diabetes, when insulin signaling and hence PI3K signaliing is impaired. On the other hand hyperactive PI3K signaling acts to drive many types of cancer. Diabetes and cancer are two main age-related diseases and interestingly deregulated insulin/ PI3K signaling towards the FOXO/DAF16 class of transcription factors also impacts lifespan or ageing of various model organisms. We are trying to understand this pivotal role of PI3K signaling in connecting age to disease and thus to understand how age contributes to disease in particular cancer.
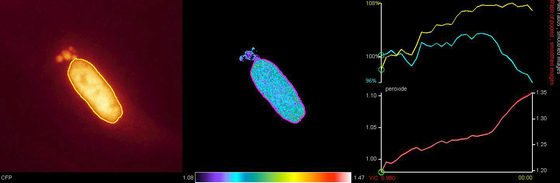
Dynamics of FOXO protein folding studied by FRET analysis. Cell expressing CFP-FOXO4-YFP was stimulated with Hydrogen peroxide to activate FOXO4 and emission of YFP/CFP was followed after CFP excitation by live imaging.
Metabolic regulation in cancer and differentiation
We study the interaction between protein signaling networks with that of metabolic pathways. To this end we make use of various model systems and at present this is predominantly the organoid culture system. Small intestinal organoids are studied amongst others at the signal cell level by using various sensors that can monitor various aspects of metabolism (see figure B). We use also all kinds of genetic perturbations in order to understand how metabolism and signaling within a single cell type contribute to the life of this cell type in the context of other cell types.
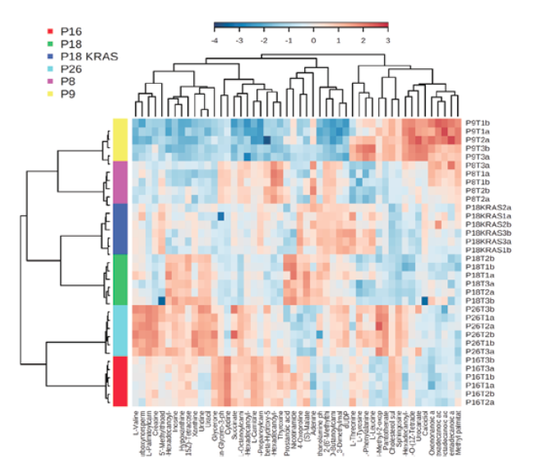
Figure illustrating the results of metabolomics analysis of patient derived colon cancer organoids by Direct Infusion mass spectrometry. Results are clustered in a heat map to indicate the existence of patient specific metabolic perturbations in these organoids.
All projects are highly technology driven and therefore we make use in house of all modern day technology including mass-spectrometry for proteomics and metabolomics, microscopy for live imaging etc.
Group members
- Ing. Lydia Smits - technician
- Ing. Miranda van Triest - technician
- Ing. Pauline Polderman - technician
- Ing. Robert van Es - technician
- Ing. Yuen Fung Tan - technician
- Richard van Schaik - technician
- Dr. Edwin Stigter - senior scientist
- Dr. HarmJan Vos - senior scientist
- Drs. Marten Hornsveld - PhD student
- Drs. Sabina van Doeselaar - PhD student
- Drs. Tianshu Gui - PhD student
Areas of expertise uitklapper, klik om te openen
Boudewijn Burgering is a Professor in Signal Transduction and head of the research section Molecular Cancer Research of the CMM at the UMCU. Boudewijn is also head of the Graduate School Cancer, Stem Cells and Development (CSnD), a visiting scientist to NCBS/InStemm Institute in Bangalore and a visiting Professor Second College of Clinical Medicine of Guangzhou, China. Boudewijn is running together with Prof Dr Nanda Verhoeven Duif the Utrecht Centre for Medical Metabolomics that provides services and expertise to researchers that wish to study metabolism within the context of their research. Also at MCR proteins@work is located, which provides support in proteomics for the UMCU.
Boudewijn is an elected member of EMBO and reviewer for the ERC, has been rewarded a VICI grant in 2002 and participates in the Dutch consortium on cancer research (CGC.nl).
The past work mainly focussed on the function of proteins within signalling networks and to decipher the molecular lay-out of signalling networks. This has led to important and highly cited discoveries e.g. PKB/AKT downstream of PI3Kinase (together with Prof. Paul Coffer). Current work has expanded towards other signalling networks most importantly the involvement of metabolism in signalling and cell fate determination but also redox-signalling in collaboration with Dr Tobias Dansen.
More contact information uitklapper, klik om te openen
Visiting address:
Stratenum 3.201
Universiteitsweg 100
3584 CG Utrecht
The Netherlands
Secretariat:
Cristina Arpesella
m.c.arpesella@umcutrecht.nl
+31 88 75 68988
Key publications uitklapper, klik om te openen
- FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. Nat Cell Biol. 2006 Oct;8(10):1064-73.
- Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, Korswagen HC. Science. 2005 May 20;308(5725):1181-4.
- Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress.Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM. Nature. 2002 Sep 19;419(6904):316-21.
- AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Medema RH, Kops GJ, Bos JL, Burgering BM. Nature. 2000 Apr 13;404(6779):782-7.
- Direct control of the Forkhead transcription factor AFX by protein kinase B. Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM.Nature.1999 Apr 15;398(6728):630-4.
- Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Burgering BM, Coffer PJ. Nature. 1995 Aug 17;376(6541):599-602.
Link to full list of publications
